Farber Lab
The Farber Lab uses innovative systems genetics and genomics approaches to study complex bone phenotypes related to physiology and disease.
Charles Farber
Associate Professor, Public Health Sciences
University of Virginia Center for Public Health Genomics
Research Focus
Common diseases, such as osteoporosis, impart significant societal health burdens. These diseases are, in part, regulated by genetic determinants and understanding their genetic basis is critical to the development of effective therapeutics. In recent years, technological advances, such as sequencing the genomes of multiple species and the ability to perform bioassays in a massively parallel fashion, have made it possible to begin to understand disease in a systems context. Systems-biology attempts to determine the direct and interactive roles of all cellular and physiological components (transcripts, proteins, metabolites, etc.) in diseased and normal states using data from high-throughput genomic studies.
Our lab is using systems approaches to investigate the molecular basis of bone strength. The goal of this work is to combine genetics and global gene expression profiling in the mouse to identify genes and pathways which influence bone strength related traits using techniques such as causality modeling between expression and physiological traits and generating disease focused gene co-expression networks. An additional component of this work is testing hypotheses generated by systems-analyses using in vitro cell based assays and transgenic mouse models.
Interests
- Bioinformatics and Genomics
- Computational Biology
- Genetics
- Metabolism
- Molecular Biology
- Single cell RNA-seq and ATAC-seq
Education
PhD in Genetics
University of California, Davis
MS Genetics
Michigan State University
BSc Biochemistry
Western Kentucky
Abdullah "Arby" Abood
PhD candidate
About Arby
Arby is a PhD candidate at the University of Virginia School of Medicine. He uses systems genetics approaches to elucidate the genetics of osteoporosis.
Interests
- Systems Genetics
- Computational Biology
- Genetic Epidemiology
Education
PhD in Molecular Genetics, 2023
University of Virginia
MSc in Micorbiology, 2018
Clemson University
BSc in Biology
George Mason University
About Atum
Atum Buo, Ph.D. is a postdoctoral fellow in the Center for Public Health Genomics. Atum specializes in bone research and his current work focuses on elucidating the impact of genetic variation on bone strength and skeletal health. Dr. Buo hopes that his research will lead to the discovery of new genes that play a role in bone strength and ultimately contribute to the development of therapies against bone diseases such as osteoporosis.
Interests
- Systems Genetics
- Bone Strength
Education
PhD in Molecular Medicine, 2017
University of Maryland
BSc in Chemistry
Haverford College, 2007
About Gina
My primary focus in the lab is development of new assay methods for the measurement of osteoblast and osteoclast phenotypes as well as the development of novel protocol methods and mouse models to meet our research needs.
Interests
- Systems genetics
- Mouse genetics
- Bone biology
Education
BSc in Molecular and Cell Biology
Penn State University
Basel Al-Barghouthi
Ph.D. candidate @ The Farber Lab
University of Virginia
About Basel
I am a Ph.D. candidate in Dr. Charles Farber’s lab at the University of Virginia. I’m currently interested in the systems genetics of complex traits, and my current research deals with unraveling the systems genetics underlying bone strength and strength-related traits.
In my research, I utilize computational tools and methods, such as transcriptomic network learning, as well as molecular biology bench work, in order to further understand the genetics underlying bone strength.
Interests
- Systems genetics
- Complex trait genetics
- Network biology
- Genomics
- Bioinformatics
Education
PhD in Biochemistry & Molecular Genetics, 2021 (ongoing)
University of Virginia School of Medicine
MS in Biological and Physical Sciences, 2017
University of Virginia School of Medicine
MS in Bioinformatics, 2015
University of Michigan
BS in Biology, 2013
The University of Texas at Austin
About Will
I am a biostatistician at the University of Virginia Center for Public Health Genomics. My research leverages system genetics approaches to elucidate the gentics of bone strength. As a computational researcher, I conduct analyses for bulk RNA-seq, single cell ATAC-seq and RNA-seq, and transcriptome wide association studies.
Interests
- Biostatistics
- Bioinformatics
- Systems Genetics
- Computational Biology
- Genetic Epidemiology
Education
MSc in Biostatistics, 2018
Northwestern University
BSc in Biology
Penn State University
About Larry
I have spent over 25 years studying varying aspects of DNA replication and genome instability, from in vitro studies of polymerase elongation complexes to the development of methods for whole-genome studies in mammalian systems. In the last 7 years I have been working on developing a pipeline for verifying candidate genes from GWAS studies using mouse models coupled with human and mouse cell model systems.
Interests
- Systems Genetics
- Biology
- Biochemistry
Education
PhD in Biochemistry, 1992
University of Virginia
BSc in Botany
University of Iowa
Projects
GWAS
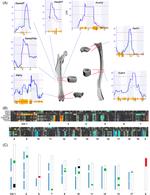
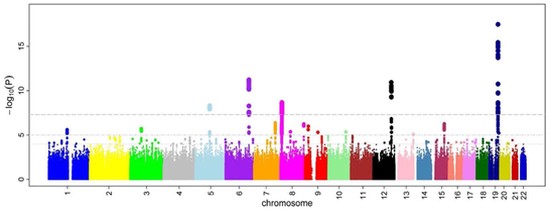
GWAS have identified dozens of loci for bone mineral density (BMD). The next step is to use these data to identify the genetic variants and genes responsible for the genetic effects.
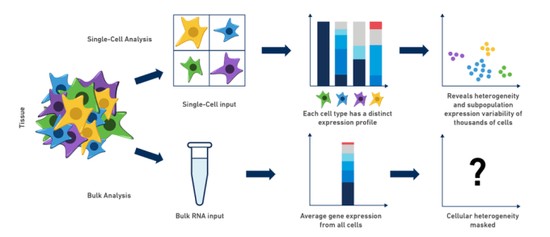
Single Cell
Skeletal development and maintenance is controlled by numerous cell-types. Historically, genomic studies of bone cells have been challenging due to difficulties in isolating homogenous cell-types from marrow or bone. Recently, this has begun to change with the emergence of single cell technologies
Featured Publications
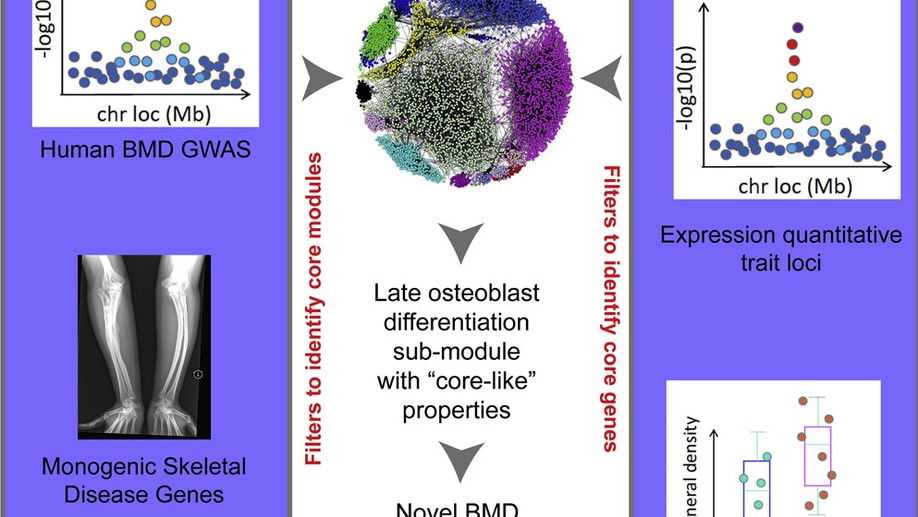
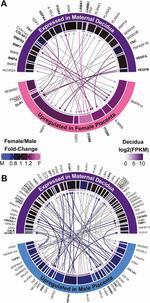
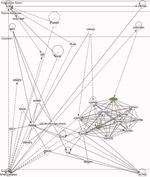
Identification of a Core Module for Bone Mineral Density through the Integration of a Co-expression Network and GWAS Data
The omnigenic model of the genetic architecture of complex traits proposed two categories of causal genes: core and peripheral. Core genes are hypothesized to directly regulate disease and may serve as therapeutic targets. Using a cell-type- and time-point-specific gene co-expression network for mineralizing osteoblasts, we identify a co-expression module enriched for genes implicated by bone mineral density (BMD) genome-wide association studies (GWASs), correlated with in vitro osteoblast mineralization and associated with skeletal phenotypes in human monogenic disease and mouse knockouts. Four genes from this module (B4GALNT3, CADM1, DOCK9, and GPR133) are located within the BMD GWAS loci with colocalizing expression quantitative trait loci (eQTL) and exhibit altered BMD in mouse knockouts, suggesting that they are causal genetic drivers of BMD in humans. Our network-based approach identifies a core module for BMD and provides a resource for expanding our understanding of the genetics of bone mass.
Recent Publications
Contact
- 434-243-8584
- OMS, Room 3817B , Charlottesville, VA 22908
- DM Me
- Discuss on Forum